- Subscribe to RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
Write to Binary File Cluster Size
Solved!10-02-2015 12:40 PM - edited 10-02-2015 12:41 PM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
I have a cluster of data I am writing to a file (all different types of numerics, and some U8 arrays). I write the cluster to the binary file with prepend array size set to false. It seems, however, that there is some additional data included (probably so LabVIEW can unflatten back to a cluster). I have proven this by unbundling every element and type casting each one, then getting string lengths for the individual elements and summing them all. The result is the correct number of bytes. But, if I flatten the cluster to string and get that length, it's 48 bytes larger and matches the file size. Am I correct in assuming that LabVIEW is adding some additional metadata for unflattening the binary file back to a cluster, and is there any way to get LabVIEW to not do this?
I'd really prefer to not have to write all 30 cluster elements individually. A different application is reading this and it is expecting the data without any extra bytes.
Solved! Go to Solution.
10-02-2015 12:44 PM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
Had overlooked this in context help:
Arrays and strings in hierarchical data types such as clusters always include size information.
Well, that's a pain.
10-02-2015 01:15 PM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
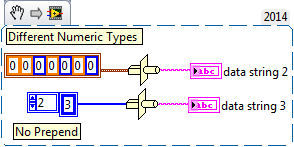
Yup I remember reading a CLAD question on this. It had a 2D array of U8, that was 2x2 and it asked if you write this to a binary file how much space does it take up? The obvious answer is 4 bytes, but the correct answer is 4 bytes plus an I32 for each dimension. No idea the work around. Maybe cast to a string and write that? Or cast to a 1D array of bytes, then write to file, then delete the first 4 bytes of the file (which would be the I32 of size). Why do you want to write to file without this information?
Unofficial Forum Rules and Guidelines
Get going with G! - LabVIEW Wiki.
17 Part Blog on Automotive CAN bus. - Hooovahh - LabVIEW Overlord
10-02-2015 02:04 PM - edited 10-02-2015 02:09 PM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
@Hooovahh wrote:
Why do you want to write to file without this information?
Because the existing parser does not expect that information there so it chokes. The workaround was just to do as follows, writing each element individually in its own case. When writing arrays that aren't within a cluster to the file and setting the prepend array size to false, it works. Of course if an element is added to the cluster, I will get a runtime error when I hit the default case, but this is a pretty fixed format so I'll cross that bridge when I come to it.

10-02-2015 02:10 PM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
You will still need to separate but you can include just the numbers in one cluster and the arrays separately. This might give you less than 30 individual writes.
mcduff
10-02-2015 03:05 PM - edited 10-02-2015 03:06 PM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
@mcduff wrote:
You will still need to separate but you can include just the numbers in one cluster and the arrays separately. This might give you less than 30 individual writes.
mcduff
Doesn't work if the array is defined to be somewhere in the file in between two scalars in the first cluster. The easiest way to do it is just write each element individually so I have control over it, eventhough it's a bit painful.
10-02-2015 03:33 PM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
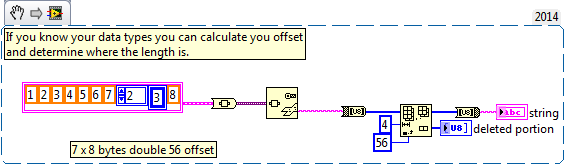
If you know your data type you can calculate the offset where the length is prepended.
Cheers,
mcduff