- Subscribe to RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
- « Previous
-
- 1
- 2
- Next »
Efficent lookup tables. To provide bit error correct to images.
08-08-2016 07:55 PM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
If possible, please post more of your code, since it's possible there's a change elsewhere that would speed things up.
08-08-2016 09:49 PM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
You should also set execution priority to "time criticial". Finally, if the image size is known up front, and the lookup tables are constant, you could probably decrease execution time by using an "in place element" structure. This prevents LabVIEW from reallocating variable size arrays as you go. On a similar note, it's best that the arrays in question are initialized with 0's and of a specific size to start with. Again, as was mentioned, these vague suggestions could be made more concrete if you share your code. If you can, do Save As > Duplicate File Hierarchy. Then zip the folder and upload it.
08-08-2016 09:51 PM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
Our data package consists of modulating a hardware state at 135 hz between 4 states, we then sum each of those states 512 times for each package.
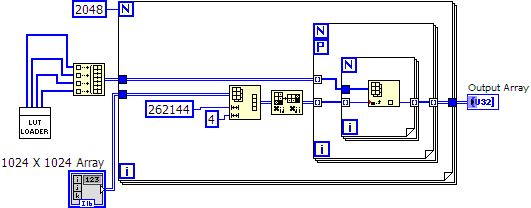
The Lookup table correct for each adc is 12 bits (4096 values). Right now I am just focused on getting the lookup correction fast. Currently it takes ~16 seconds to do 2048 corrections on a test array of 1024x1024 random nubmers. The whole data processing task (including a lot of other steps) for 2048 images needs to be under 15.15 seconds. So I think I need to find a factor or ~2 increase in speed.
Thanks
08-08-2016 10:24 PM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
What exactly is the point of the outer For loop? It runs 2048 times, but each time it runs the data within is discarded.
08-08-2016 10:28 PM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
08-09-2016 01:59 AM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
Parallellize the outmost loop (and only that loop), it's generally the most efficient. The Array subset and transform can also be done outside the loops.
/Y
08-09-2016 04:11 AM - edited 08-09-2016 04:24 AM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
If you can re-use the output data from the last iteration as the basis for the next iteration then you can shave some time off the memory allocations.
Without enabling parallel loops I have a version which does each 1024x1024 image in approximately 6.2 ms on my machine (just over 12s for 0248 images).
I tried enabling parallel loops but the memory arbitration which then occurs significantly degrades performance. It's much faster to work through the array from 0..N due to how RAM works.
Would it be possible to process, say 4 images in parallel, each image working serially? That should get you down to around 4-5 seconds per 2048 images.
Shane.
Hmm, I notice something strange when performing calculations on 4 cores simultaneously (4 images concurrently). VI Profiler shows 26s for one iteration, but the software is running clearly faster than that, it's updating in approximately 7 seconds (for 2048 images). Seems like the VI Profiler has trouble benchmarking code over more than one core. Maybe this is not news, but I did not know the profiler worked like that.
08-09-2016 05:53 AM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
I reckon NathanDs method is the best for a single Image, but I would still recommend parallelising subsequent images rather than parallelising the operations within a single image. By splitting the images over 4x VIs like NathanD has written, I get the 2048 images processed in 3.3s.
Shane
- « Previous
-
- 1
- 2
- Next »