- Subscribe to RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
efficiently time average a lot of data
09-22-2015 03:40 PM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
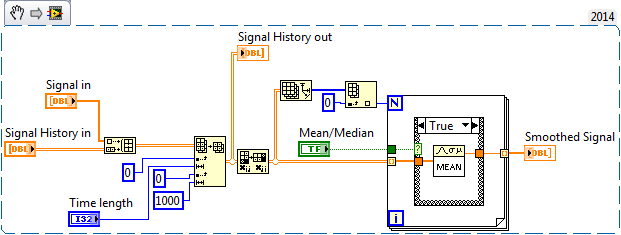
Hi. I am wanting to time average (median or mean) a lot of data efficiently and I am struggling with how to do it. Below is a snippet of the code I have been using. The code works, but it doesn't scale well. Specifically, in my application, a Time Length of about 100 really starts to bog down. My signals lengths are fixed at 1000 and they are generated at 20 Hz. I'm sure there is a more appropriate way to go about this. Can someone suggest something?
09-22-2015 03:47 PM - edited 09-22-2015 03:49 PM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
20Hz isn't very fast, so this shouldn't be too much of an issue.
Some developer notes:
- All of your zero indices can be deleted since the array operations default to zero.
- You don't need to pull the array size, etc for the For loop. The auto-indexing takes care of the iteration count for you, so you can leave N unwired.
- You could try configuring parallelism on your For loop to see if that speeds it up more for you. (Right click)
What are some standard array sizes for those inputs? What is the unit of Time Length? It's indexing the array, so is it in units of 1/5th of a second?
Thanks for attaching the snippet instead of a screenshot. ![]()
Cheers
--------, Unofficial Forum Rules and Guidelines ,--------
'--- >The shortest distance between two nodes is a straight wire> ---'
09-22-2015 07:54 PM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
I am awawre of points 1 and 2, and 3. I wired the indicies and for loop limit just to be explicit for other people who aren't as familiar with LabVIEW. I haven't even considered using a parallel for loop for this routine, I use them elsewhere in my code.
Typical array sizes are 1000xN where N is 20-200 (1-10s for 20 Hz data). The 1000 is fixed. N=20 works fine. N=200 is when things start to bog down. I should also note that this routine runs on 8 channels of synchronized data, so 8 1000xN arrays.
09-22-2015 08:28 PM - edited 09-22-2015 08:41 PM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
I wasn't talking about parallel For loops, I was talking about parallelizing your For loop. When you right click it, you can set that up and set the number of parallel instances to match your number of cores. If you're running the VI in parallel with itself 8 times already, this might not help much.
I'm curious what you mean by bog down because even 1000x200 isn't toooo bad.
Cheers
--------, Unofficial Forum Rules and Guidelines ,--------
'--- >The shortest distance between two nodes is a straight wire> ---'
09-22-2015 08:40 PM - edited 09-22-2015 08:52 PM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
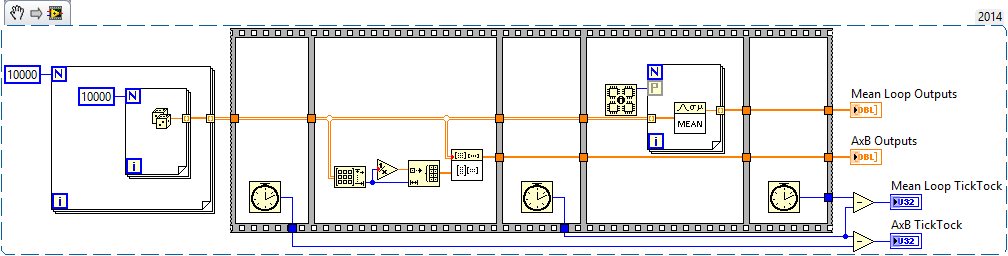
Never fear! There is another way. Math to the rescue:
Check out this benchmark I made for comparing a matrix operation against the Mean loop. You'll find the AxB to be much faster.
I set up the parallelization on the For loop, but changed it to debugging mode to disable the parallelism for the benchmark. If you turn on the parallelism again you'll see it's faster, but like I said above if you're already doing this processing in parallel it won't help much.
Cheers
--------, Unofficial Forum Rules and Guidelines ,--------
'--- >The shortest distance between two nodes is a straight wire> ---'
09-23-2015 08:02 AM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
Thanks for the code snippet. It makes sense that calculating the mean via matrix multiplication is faster than using LabVIEW level for loops. The matrix multiplication version likely spends most of its time in the linear algebra subroutines and the for loop version probably goes in and out of them with each loop iteration. I will modifiy my mean code to see how this effects my overall application.
The big issue I still face is the calculating the median. In my original code snippet, the true case calculates the mean while the false calculates the median. Calculating the median is slower than the mean due to sorting.
09-23-2015 09:24 AM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
In any (ANY) scenario like this, you need to be aware of what causes what.
You should carefully measure, using a tool like this, the time for each stage of your process.
A rule I go by is this: You can NOT make the computer go faster. You CAN make it do less work.
You might find (I'm not sure) that it's FASTER to keep your data organized the way it is (avoid the TRANSPOSE operation) and pick out the data yourself.
Or simply organize it differently to start with: instead of NSamples x NChannels, keep it NChans x NSamples all the way.
You should not just assume that the MEAN /median function is the bottleneck.
MEAN is simple enough to do yourself, simply inline a SUM ARRAY ELEMENTS with an ARRAY LENGTH and a DIVIDE operation - that avoids a VI call.
ANYTHING you can do to avoid shuffling memory around will get you improved speed.
Culverson Software - Elegant software that is a pleasure to use.
Culverson.com
Blog for (mostly LabVIEW) programmers: Tips And Tricks
09-23-2015 10:22 AM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
Do you need full history?
If yes, and size is not defined, I would extend it in big portions, then replace single row with new data.
Do you change Time length during run-time?
It might be efficient to keep "old average", subtract the eldest data, add new data. Why add intermediate data every time?
09-23-2015 11:01 AM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
Wouldn't you be able to use the ptbypt mean?
Typically it is much more efficient to sum the data immediately and keep track of the sum and the number of points. To get the mean at any time, simply divide the two. It is much more efficient to keep a few scalars than piles of large arrays.
09-23-2015 12:06 PM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
The mean isn't the big time killer, it is the median (due to sorting). How do you run a point-by-point VI on each element of an array if the poin-by-point VIs are reentrant?